

In Bayesian inference, a posterior probability is derived from a prior probability and a likelihood function. I-PINE adopts this idea to provide robust and reliable chemical shift assignments. After generation of spin systems, connectivities between spin systems are infered by analyzing different NMR experiments. Knowledges acquired from different conditions are connected by Belief propagation. To obtain spin system assignment, connectivities of CA, CB and CO are calculated, and secondary structure prediction (PECAN) and linear analysis of secondary chemical shift (LACS) follow. These steps reiterates to maximize the results. For side chain assignments, initial probability density functions for all available atoms are generated from filtered-BMRB statistical histogram at first. Bayesian updates on the probability density functions follow by analyses of different NMR experiments, and peaky probabilities indicating probable assignments for the atom are introduced. Then, factor graph analyses using peaky probabilities for all available atoms in each residue are iteratively applied.
In I-PINE, the PACSY DB has been aplied to the bayesian framework. As a result, overall quality and quantity of assignments have improved. Also, more experiments are supported. High contrast probability curve has been achived that means more accurate chemical shifts given with high probability and less accurate chemical shifts given with low probability. Indeed, user can rely more on probability from I-PINE. Initial secondary structure probabilities for helix (H), strand (E) and coil (C) are now predicted by GetSBY (Get Secondary structures By PACSY), which were used to be fixed values for all residues (H: 0.327; E: 0.291; C: 0.382). Initial probability density functions are now combination of filtered-BMRB statistical histograms and PACSY-derived gaussian functions. PACSY chemical shift analysis of different size peptides (7-mer, 5-mer, 3-mer, 2-mer) are being conducted on-the-fly to refine initial probability density functions that results significant enhancement of sidechain assignment.
Simply, the way I-PINE web server operates is a) to accept user inputs from web page, b) to run the I-PINE algorithm, and c) to send results to the user. To establish this framework, we employed various computer techniques in our servers. A figure below illustrates how I-PINE web server is designed and what techniques are employed.
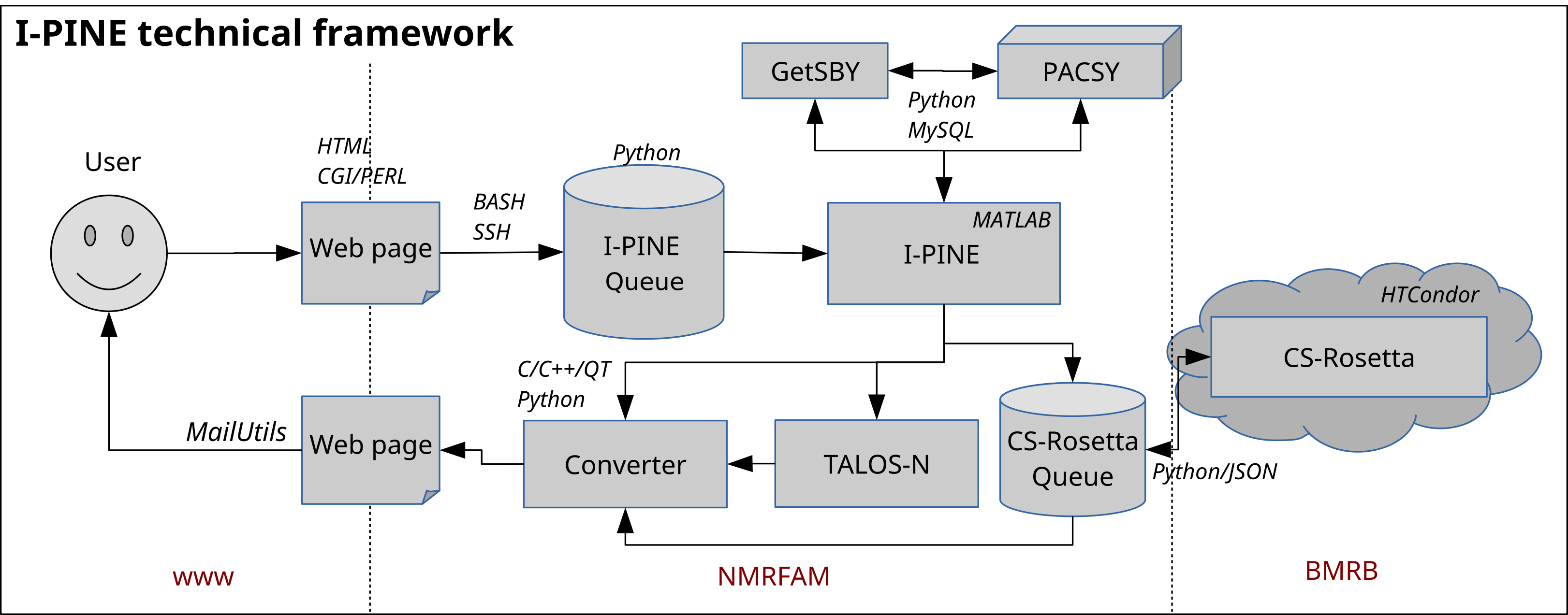
Upon agreement, we fix bugs and support users. We encourage users contacting us for any issues because it improves our development and results better quality of outcomes that benefits users eventually. Data are accessible by using provided job identification key provided by a user or the one included in the result email and the computer screen right after the submission. Only the numbers of submissions over the years are used to find growths of I-PINE usage in presentations and grant submissions.
The Integrative NMR is an NMR platform that makes biomolecular NMR spectroscopy much more accessible by integrating tools, databases, web services, and video tutorials that can be launched by simple installation of NMRFAM software packages or using a cross-platform virtual machine that can be run on any standard laptop or desktop computer. Resonance assignment in the platform is fundamental ground that can extend to any other biomolecular NMR researches, but it is very time consuming process. Therefore, I-PINE is one of the most important modules in the platform. It automates the time consuming process by combining computational approaches, AI approaches and NMR theory. Please read our paper to know what other things are doable.